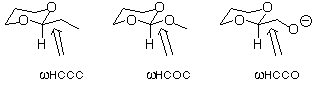
| Chemistry 8003 | Computational Chemistry | 4 Credits | Winter Quarter 1999 | ( Due 1 / 25 / 99 ) |
Using PC Model, answer any two of the three questions
below.
1. PCModel includes a number of substructure menus designed to
facilitate the building of biopolymers. From the amino acid menu,
construct the oligopeptide lys-lys-lys. In a biological system
the C terminus would be deprotonated and the N terminus protonated,
but let's not worry about that. The objective of this problem
is to find the lowest energy conformation you can for this tripeptide.
Presumably this will involve some internal hydrogen bonds. Grading
will be based on the range of energies found (lower is better).
When you have found what you think to be the lowest energy structure,
print it out using the print option that includes the energy-try
to orient your structure and choose the display option (e.g.,
ball-and-stick, tube, etc.) so as best to illustrate the actual
conformation. [Note: On the Macintosh, some versions of PCModel
will not send the PostScript file to the printer until you bring
the Finder to the forefront, e.g., by touching the desktop.]
2. Use the dihedral driver option in PCModel to construct torsional
potentials about the indicated bonds in the three molecules below.
Construct the potentials over the range of 180 degrees taking
points every 15 degrees (symmetry dictates that only 180 degrees
are unique). Note that PCModel saves the 12 structures to a file,
with their associated energies, so you can open any one of them
to look at its energy and get the breakdown of that energy into
its force field components by doing a single point. By analyzing
the contributions of different force field terms to the total
energy, provide a chemical explanation for any important differences
you perceive between the three potentials.
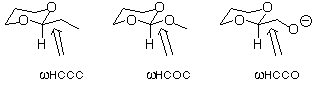
3. Design a problem of your own that uses PCModel to illustrate
some chemically interesting concept. Write down the problem, and
then provide the answer. Note that you do not need to pick something
that PCModel performs well for! However, if you create a problem
where PCModel clearly gives the wrong prediction, provide some
discussion in your answer of why the force field fails to be accurate.
To get more of a feel for typical problems, feel free to drop
by the website and look at first problem sets from previous years.
Grading will be based on quality of the problem and originality.
Important note on resources: In addition to the computers
in 176 Kolthoff, there are platforms in the IT Labs that are running
PC Model. In principle, you have access to these machines
(assuming you've been assessed the IT computer fee-if you have
any trouble, let me know). Locations are EE/CSci 3-170 (M-Th 7
am - 2 am; Fr 7 am - midnight; Sa-Su 10 am - 2 am) and Physics
130 (M-Th 8 am - 10 pm; Fr 8 am - 6 pm; Sa 10 am - 6 pm; Su 4
pm - 10 pm). For output you will need a printer access card, which
can be obtained in the lab itself.