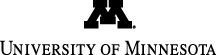
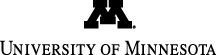
Main navigation | Main content
Recent research from Professor Jiali Gao's group
Protein dynamics are essential to the catalytic function of enzymes. However, the precise mechanism by which large-scale motions and fast, local fluctuations are connected to the chemical transformation to lower the free energy barrier remains poorly understood and is a central question in current enzymology. In a recent paper, published in Biochemistry (2013, 52, 2036-2049) as part of a special Current Topic selection on enzyme mechanisms, graduate student Yao Fan from the Department of Biochemistry, Molecular Biology and Biophysics, and postdoctoral associates Alessandro Cembran and Shuhua Ma in Professor Jiali Gao's group reported the finding that distant double mutations (M42W and G121V), ca. 15 angstroms away from the active site in dihydrofolate reductase (DHFR), significantly reduce the dynamic motions of a flexible loop, called M20 loop, at the transition state of the catalyzed hydride transfer. The computational results helped to interpret the experimental observations of increased entropic barrier, reduced catalytic activity, and altered kinetic behaviors as revealed by temperature-dependence of kinetic isotope effects (KIE). Importantly, it provided direct, albeit computational, evidence that the diminished rate enhancement in the mutant enzyme is a result of protein dynamics knock-out, an idea that was recently explored experimentally on other mutations in DHFR (G. Bhabha, et al., Science, 2011, 332, 234). A mechanism, extending the "ensemble" view of protein allostery to enzyme catalysis, was proposed to rationalize the computational and experimental findings.
The computational studies were possible thanks to several methods developed in the Gao group, including combined quantum mechanical and molecular mechanical (QM/MM) techniques, and an integrated path integral-free energy perturbation and umbrella sampling (PI-FEP/UM) approach for accurately determine primary and secondary KIE with a precision comparable to experiments.
Figures:
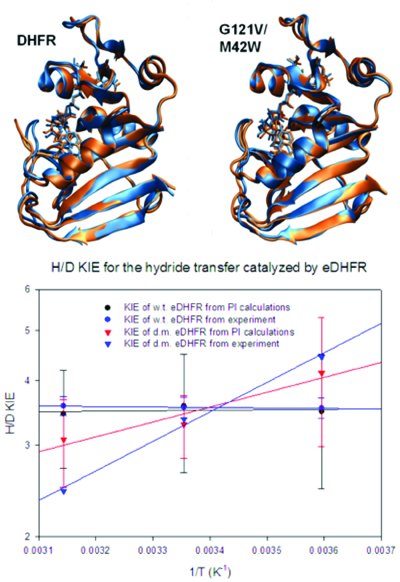
(Top) Conformational dynamics of the M20 loop that undergoes a closed to open transition from the Michaelis complex (blue) to the transition state (brown) in wild-type DHFR (left). However, the M20 loop is locked in the closed conformation, a dynamics knock-out, throughout the chemical step in the M42W/G121V double mutant (right).
(Bottom) The altered loop dynamics by distant mutations is reflected kinetically in the temperature-dependence of measured and computed kinetic isotope effects.